SpatialOmics in Life Science
Protein analysis or whole transcriptome expression in user-defined specific regions of tissue sections.
Background
Combined understanding of the cellular organization and functionality of individual cells in tissue will lead to advances in basic understanding but also improved strategies to target various diseases.
The possibility to perform deep molecular profiling of specific cell types or spatially defined areas of imaged tissue, open up for investigations on the functional impact of tissue heterogeneity, and when relevant, connect it to patient outcome.
Multiplex analyses of large patient cohorts have for a long time been restricted to bulk samples and limited by the availability of sample formats feasible for global/high-plex molecular analysis. Recent parallel technological development within the field allows the use of diverse format of intact tissue and next generation sequencing of large barcode-based panels as briefly described below.
Our technologies
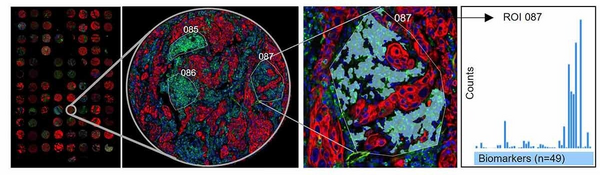
GeoMx Digital Spatial Profiler
The GeoMx Digital Spatial Profiler is designed for highly multiplexed, spatially resolved expression analysis of protein or mRNA targets in tissue sections from FFPE or fresh frozen samples. Analyze up to 570 protein or 18,000 mRNA targets on each tissue slide and in each ROI. The GeoMx can handle slides with whole tissue sections, multiple sections per slide as well as tissue microarrays. There are targeted panels for immuno-oncology, immunology and neurobiology as well as whole transcriptome panels for human and mice tissue.
Tissue sections are stained with fluorescently labelled markers and DSP probes for protein and/or RNA detection. The slides are scanned and regions of interest are selected based on the imaged markers. Each ROI can be segmented to select specific cell types. One at a time, each ROI is exposed to UV-light and the cleaved off target specific barcodes are collected separately. When all ROIs are collected the barcodes in each ROI are quantified and mapped back to the tissue.
nCounter platform
The nCounter system is developed for multiplex gene expression analysis in total RNA of up to 800 gene targets per reaction. It is suitable for identification of disease specific biomarkers, verifying gene signatures or investigating specific biological pathways.
There are a range of available panels targeting disease or biological pathways:
Oncology and Immuno-oncology
Autoimmune diseases and response
Immunology, Inflammation and host responses to infection
Neurobiology and Neuroinflammation
miRNAs
Publications
Anders Valind, Bronte Manouk Verhoeven, Jens Enoksson, Jenny Karlsson, Gustav Christensson, Adriana Mañas, Kristina Aaltonen, Caroline Jansson, Daniel Bexell, Ninib Baryawno, David Gisselsson & Catharina Hagerling. Macrophage infiltration promotes regrowth in MYCN-amplified neuroblastoma after chemotherapy. OncoImmunology 2023 12:1, DOI: 10.1080/2162402X.2023.2184130
Sobti A, Sakellariou C, Nilsson JS, Askmyr D, Greiff L, Lindstedt M. Exploring Spatial Heterogeneity of Immune Cells in Nasopharyngeal Cancer. Cancers. 2023; 15(7):2165. https://doi.org/10.3390/cancers15072165
Anna Sandström Gerdtsson, Mattis Knulst, Johan Botling, Artur Mezheyeuski, Patrick Micke & Sara Ek Phenotypic characterization of spatial immune infiltration niches in non-small cell lung cancer. OncoImmunology, 2023 12:1, DOI: 10.1080/2162402X.2023.2206725
